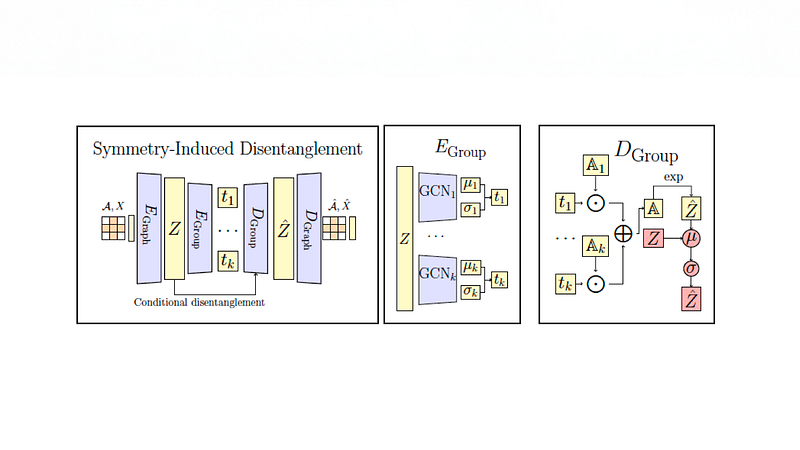
Generative models for molecules are hard to interpret – their latent spaces are typically overly convoluted. Learning disentangled representations is important for unraveling the underlying
complex interactions between latent generative factors pertaining to different molecular properties of interest (e.g., ADMET properties). For example, we might want to improve the metabolism property (‘M’ in ADMET) of a molecule without significantly altering its other properties.
However, not all properties can be disentangled – e.g., usually it’s hard to optimize or improve bioactivity of a candidate molecule without enhancing its toxicity as well. In a NeurIPS 2022 paper, we address this issue with a novel concept of conditional disentanglement: our method segregates the latent space into uncoupled and entangled parts. This allows us to exercise finer control over molecule generation and optimization. Experimental results on molecular data strongly corroborate the interpretability of our method. Interestingly, this interpretability is accompanied with improved molecular generation performance, as evaluated across multiple standard metrics.